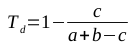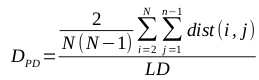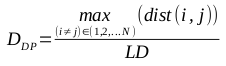I have recently been working a lot with OTU tables in the BIOM (Biological Observation Matrix) format, generated by QIIME and PIPITS. This now-common OTU table format seems to be getting more popular; I don’t recall running into such files in my bioinformatics studies just a few years back.
Tool suites like QIIME allow you to treat BIOM-formatted OTU tables as black boxes, keeping the users insulated from their internals, but I wanted to examine them a bit in Python. They are HDF-formatted files, version 5 in recent tools. Installing the h5py package along with the ancillary tables package seems to be a good way to go, but does not work. The tables package does not handle variable length strings.
The easiest way to inspect OTU tables in Python is with the biom-format package, which interestingly uses the h5py package that I could not get to work.
For starters, installation:
pip install biom-format pip install h5py pip install pandas pip install numpy
And now a tour through a sample OTU table. The biom-format documentation didn’t make it obvious how to open an BIOM file, but it turns out that they provide a load_table routine that accepts a filename. There is a ‘sample’ axis and an ‘observation’ axis, where the observations are the OTUs.
>>> from biom import load_table
>>> t = load_table(‘sample_otu_table.biom')
>>> print(t.ids())
['SA8' 'Bed' '26' '29' 'Food' 'CA4' '5' 'SA3' 'Water' '15' '22' 'SA15'
'SA25' 'CA22' '1' 'TRI3' '24' '10' '14' 'PA36' '3' '7' '16' '21' 'PA17'
'CA8' '25' 'PA60' 'PA29' 'PA71' 'PA53' 'PA79' 'PA64' 'PA46' 'PA6' 'TRI18'
'TRI8' '8' 'CA17' 'PA1' '13' '27' '11' '28' '9' '4' '2' '18' '12' '23'
'PA19' 'HydroGel' '6' '20' '30' '19' 'TRI24' '17']
>>> print(t.ids(axis='observation'))
['4371191' '4479944' '816470' ... '512148' '1109141' '3275744']
>>> print(t.data('SA8', axis='sample'))
[2. 0. 0. ... 0. 0. 0.]
>>> print(t.data('4371191', axis='observation'))
[2. 5. 1. 1. 1. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.
0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.
0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]
The taxonomy metadata annotations are easily available, allowing one to quickly access a list of all taxa present in the study.
>>> print(t.metadata('4371191', axis='observation')['taxonomy'])
['k__Bacteria', 'p__Proteobacteria', 'c__Gammaproteobacteria', 'o__Pseudomonadales', 'f__Pseudomonadaceae', 'g__Pseudomonas', 's__']
>>> metadata_df = t.metadata_to_dataframe('observation')
>>> metadata_df.columns = ['kingdom', 'phylum', 'class', 'order', 'family', 'genus', 'species']
>>> print(metadata_df.head())
kingdom phylum class \
4371191 k__Bacteria p__Proteobacteria c__Gammaproteobacteria
4479944 k__Bacteria p__Actinobacteria c__MB-A2-108
816470 k__Bacteria p__Firmicutes c__Bacilli
842143 k__Bacteria p__Actinobacteria c__Actinobacteria
99682 k__Bacteria p__Proteobacteria c__Gammaproteobacteria
order family genus species
4371191 o__Pseudomonadales f__Pseudomonadaceae g__Pseudomonas s__
4479944 o__0319-7L14 f__ g__ s__
816470 o__Bacillales f__Bacillaceae g__ s__
842143 o__Actinomycetales f__Cellulomonadaceae g__Cellulomonas s__
99682 o__Pseudomonadales f__Pseudomonadaceae g__Pseudomonas s__
Methods are provided to collapse the table over particular levels of taxonomy, to normalize the data across samples to account for difference in sampling depth, and many other useful operations. (This code was borrowed from the biom-format documentation.)
>>> phylum_idx = 1 >>> collapse_f = lambda id_, md: '; '.join(md['taxonomy'][:phylum_idx + 1]) >>> collapsed = t.collapse(collapse_f, axis='observation') >>> print(collapsed.head()) # Constructed from biom file #OTU ID SA8 Bed 26 29 Food k__Bacteria; p__Proteobacteria 11.17291857273559 2.9359560841720036 0.5507776761207686 0.16468435498627632 6.343092406221409 k__Bacteria; p__Actinobacteria 0.412751677852349 102.79362416107382 3.9077181208053693 1.354026845637584 0.06040268456375839 k__Bacteria; p__Firmicutes 17.308791684711995 0.4538761368557817 2.1459506279774794 1.0415764400173235 0.11173668254655696 k__Bacteria; p__Verrucomicrobia 0.0 1.5 0.0 0.0 0.0 k__Bacteria; p__Bacteroidetes 0.006493506493506494 1.7402597402597402 0.21428571428571427 0.0 0.07792207792207792 >>> normed = t.norm(axis='sample', inplace=False) >>> print(normed.head()) # Constructed from biom file #OTU ID SA8 Bed 26 29 Food 4371191 3.813009990086174e-05 7.30823199251637e-05 9.994003597841295e-05 0.0002801905295601009 1.965177062453327e-05 4479944 0.0 2.9232927970065482e-05 0.0 0.0 0.0 816470 0.0 0.0 0.0 0.0 0.0 842143 0.0 0.00011693171188026193 0.0 0.0 0.0 99682 1.906504995043087e-05 0.0 0.0 0.0 0.0