I have recently run across a few examples of classifiers in widely-used bioinformatics applications. Assessing the ability of a classifier to provide good predictions is a difficult problem. In this post and the next, I will describe the use of metrics in these applications, and how they might be improved.
Confusion Matrices
We will start with binary classification, in which samples are divided into two categories. If we are testing individuals for the presence or absence of a disease, we can divide the samples into sets of individuals who have the disease (positives) and those that do not (negatives). The classifier will attempt to divide the samples into the correct sets based on the results of testing. Those who have the disease and are correctly identified as such by the classifier are true positives (TPs). Those that do not have the disease and are correctly classified as such are known as true negatives (TNs). Misclassifications are called false negatives (FNs) for subjects that have the disease but are classified as healthy, or false positives (FPs) for subjects that do not have the disease but are classified as diseased.
These can be arranged into a confusion matrix, as in the following:
It is common to use some of the following simple metrics, all based on the contents of the confusion matrix:
- precision, the fraction of positive results that are correct: TPs / (TPs + FPs)
- sensitivity (also called recall), the faction of positive subjects that are identified as positive: TPs / (TPs + FNs)
- specificity, the fraction of negative subjects that are identified as negative: TNs / (FPs + TNs)
- accuracy, the ratio of correct classifications to the total number of samples: TPs + TNs / (TPs + TNs + FPs + FNs)
The relative importance of these metrics depends on the problem. Is it useful to sacrifice specificity for sensitivity? Is precision or specificity a better metric? Are there useful metrics for directly comparing classifiers with a single number?
Case Study: Protein Domain Prediction
The prediction of protein domains based solely on the protein’s amino acid sequence is an important technique for functional annotation. PROSITE, a database of protein families and domains on ExPASy, makes use of two types of approaches for detecting the presence of domains: patterns and profiles. Each of the approaches is essentially an example of binary classification: a protein sequence is either predicted to have instances (one or more) of a particular domain, or it is predicted to have no instances of that domain.
In PROSITE, these classifiers are assessed by their precision and recall. These metrics are widely used in information retrieval to assess the ability of a system to provide a list of relevant documents. It is common to use an F-measure to summarize the two metrics by producing a harmonic mean of their values, but this score is not used in PROSITE. Looking back at the equations for producing these values, it should be clear that the number of TNs is completely ignored; the ability of the system to correctly assess negatives is not considered. Is this important?
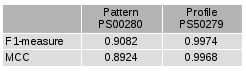
We will look at Kunitz domains, a type of protein domain, as an illustrative example. PROSITE includes two models for recognizing Kunitz domains: a pattern (PS00280) and a profile (PS50279). It is not obvious how many samples were in the overall set. At the time of this writing, the models were assessed by their ability to classify the proteins in UniProtKB/SwissProt 2013_09, which contains 540958 samples. This results in the following confusion matrices for the two classifiers:
The profile is clearly superior from a cursory glance, and this is reflected by the precision and recall metrics provided in PROSITE. For the pattern, the precision is 0.9842 and the recall is 0.8432. For the profile, the precision is 1.0 and the recall is 0.9948.
Accuracy is a Poor Metric
Accuracy is generally a poor metric, particularly when the samples are unbalanced (if, for example, there are far more negatives than positives, as is the case in this scenario). The accuracy for the pattern is 0.9998835, and for the profile is 0.9999963. It was wise to not provide accuracy scores, as they do not differentiate between the two classifiers very well.
Let’s consider what would have happened if there were far fewer TNs in the set of samples, which might have been the case if we filtered the input to not consider proteins that were very unlikely to include Kunitz domains. If there were a total of 500 proteins considered, we would have ended up with 186 TNs for the pattern, and 189 TNs for the profile. The accuracy would be 0.874 for the pattern, and 0.996 for the profile; the precision and recall metrics would remain unchanged.
Single Scores for Comparing Classifiers
While the precision and metric scores are useful, it can be difficult to compare classifiers using them. Is it more important to ensure that most positive results are correct, that most positive subjects are identified as positive, or that most negative subjects are identified as negative? Summarizing the performance of a classifier with a single number is a very useful feature.
- F-scores are measures that combine both precision and recall into a single number. The F1 measure is the harmonic mean of these two numbers: (2 * precision * recall) / (precision + recall). This formula can be modified to weigh these two numbers differently depending on their relative importance. The result ranges from 1 (best case) down to 0 (worst case).
- The Matthews correlation coefficient (MCC) can be calculated from the confusion matrix using this formula: ((TPs * TNs) – (FPs * FNs)) / sqrt((TPs + FPs) * (TPs + FNs) * (TNs + FPs) * (TNs + FNs)). This produces a number that indicates how well the predictions correlate to the actual observations. The number ranges from 1 (best case) down to -1 (worst case), with a result of 0 indicating that the classifier performs no better than random guessing.
Of course, some amount of nuance is unavoidably lost when distilling the results of a classifier down to a single number. However, such metrics can be extremely useful when comparing classifiers, and that appears to hold true for protein domain prediction.
Other Thoughts
It is notable that both the original metrics and my suggested metrics suffer from a more fundamental flaw. It is common for a protein sequence to have multiple instances of a domain; if a profile or pattern recognizes one or more such domains without recognizing them all, it is still considered a “success.” Thus, the metrics are really assessing how successful the classifiers are at establishing whether one or more domains exist in a protein, rather than how successful they are at identifying the location of every domain. In the case of Kunitz domain prediction, the pattern results in 18 partial matches, and the profile results in 17 partial matches.